Monday, December 25, 2017
Christmas tree networks
Greetings for the season.
Mathematicians live in a world of their own — individually, as well as collectively. Therefore, it is inevitable that among all of the graphs with names like pancake graphs, butterfly networks, star graphs, spider web networks, and brother trees, there should be a thing called a Christmas tree.
It was introduced by Chun-Nan Hung, Lih-Hsing Hsu and Ting-Yi Sung (1999) Christmas tree: a versatile 1-fault-tolerant design for token rings. Information Processing Letters 72: 55-63.
As you can see, it is a network, as well as a tree. The authors describe it as "a 3-regular, planar, [optimal] 1-Hamiltonian, and Hamiltonian-connected graph." The Hamiltonian characteristic refers to the existence of a path through the network that connects all of the nodes and ends up back at the start node (ie. a cycle). The Christmas tree is created by joining two Slim trees together, believe it or not. (The Fat tree is a Slim tree with an extra edge connecting the left and right nodes.)
The authors' particular interest was in communications networks (eg. computer networks); but to me it looks like a (historical) phylogenetic tree at the top with an extra network added at the bottom showing (contemporary) ecological connections. It could thus summarize all of the biology in one diagram!
You can read about all of the above-mentioned graph types in the book by Lih-Hsing Hsu and Cheng-Kuan Lin (2009) Graph Theory and Interconnection Networks; CRC Press. You need to be a mathematician to make sense of it, though.
Thanks to Bradly Alicea for alerting me to this graph type.
PS. The above diagram is actually take from: Jeng-Jung Wang, Chun-Nan Hung, Jimmy JM Tan, Lih-Hsing Hsu, Ting-Yi Sung (2000) Construction schemes for fault-tolerant Hamiltonian graphs. Networks 35: 233-245.
Labels:
Christmas
Tuesday, December 19, 2017
The art of doing science: alignments in historical linguistics
In the past two years, during which I have been writing for this blog, I have often tried to emphasize the importance of alignments in historical linguistics — alignment involves explicit decisions about which characters / states are cognate (and can thus be aligned in a data table). I have also often mentioned that explicit alignments are still rarely used in the field.
To some degree, this situation is frustrating, since it seems so obvious that scholars align data in their head, for example, whenever they write etymological dictionaries and label parts of a word as irregular, not fulfilling their expectations when assuming regular sound change (in the sense in which I have described it before). It is also obvious that linguists have been trying to use alignments before (even before biologists, as I tried to show in this earlier post), but for some reason, they never became systematized.
As an example for the complexity of alignment analyses in historical linguistics, consider the following figure, which depicts both an early version of an alignment (following Dixon and Kroeber 1919), and a "modern" version of the dame data. For the latter, I used the EDICTOR (http://edictor.digling.org), a software tool that I have been developing during recent years, and which helps linguists to edit alignments in a consistent way (List 2017). The old version on the left has been modified in such a way that it becomes clearer what kind of information the authors tried to convey (for the original, see my older post), while the EDICTOR version contains some markup that is important for linguistics, which I will discuss in more detail below.
 |
| Figure 1: Alignments from Dixon and Kroeber (1919) in two flavors |
If we carefully inspect the first alignment, it becomes evident that the scholars did not align the data sound by sound, but rather morpheme by morpheme. Morphemes are those parts in words that are supposed to bear a clear-cut meaning, even when taken in isolation, or when abstracting from multiple words. The plural-ending -s in English, for example, is a morpheme that has the function to indicate the plural (compare horse vs. horses, etc.). In order to save space, the authors used abbreviations for the language group names and the names for the languages themselves.
The authors have further tried to save space by listing identical words only once, but putting two entries, separated by a comma, in the column that I have labelled "varieties". If you further compare the entries for NW (=North-Western Maidu) and NE/S (=North-Eastern Maidu and Southern Maidu), you can see that the first entry has been swapped: the tsi’ in tsi’-bi in NW is obviously better compared with the tsi in NE/S bi-tsi rather than comparing bi in NE with tsi in NE/S. This could be a typographical error, of course, but I think it is more likely that the authors did not quite know how to handle swapped instances in their alignment.
In the EDICTOR representation of the alignment, I have tried to align the sounds in addition to aligning the morphemes. My approach here is rather crude. In order to show which sounds most likely share a common origin, I extracted all homologous morphemes, aligned them in such a way that they occur in the same column, and then stripped off the remaining sounds by putting a check-mark in the IGNORE column on the bottom of the EDICTOR representation. When further analyzing these sound correspondences with some software, like the LingPy library (List et al. 2017), all sounds that occur in the IGNORE column will be ignored. Correspondences will then only be calculated for the core part of this alignment, namely the two columns that are left over, in the center of the alignment.
In many cases, this treatment of sound correspondences and homologous words in alignments is sufficient, and also justified. If we want to compare the homologous (cognate) parts across words in different languages, we can't align the words entirely. Consider, for example, the German verb gehen
[geːən] and its
English counterpart go [gɔu]. German regularly adds the infinitive ending
-en to each verb, but English has long ago dropped all endings on verbs apart
from the -s in the third person singular (compare go vs. goes). Comparing
the whole of the verbs would force us to insert gaps for the verb ending
in German, which would be linguistically not meaningful, as those have not been
"gapped" in English, but lost in a morphological process by which all endings
of English verbs were lost.There are, however, also cases that are more complicated to model, especially when dealing with instances of partial cognacy (or partial homology). Compare, for example, the following alignment for words for bark (of a tree) in several dialects of the Bai language, a Sino-Tibetan language spoken in China, whose affiliation with other Sino-Tibetan languages is still unclear (data taken from Wang 2006).
 |
| Figure 2: Alignment for words for "bark" in Bai dialects |
In this example, the superscript numbers represent tones, and they are placed at the end of each syllable. Each syllable in these languages usually also represents a morpheme in the sense mentioned above. That means, that each of the words is a compound of two original meanings. Comparison with other words in the languages reveals that most dialects, apart from Mazhelong, express bark as tree-skin, which is a very well-known expression that we can find in many languages of the world. If we want to analyze those words in alignments, we could follow the same strategy as shown above, and just decide for one core part of the words (probably the skin part) and ignore the rest. However, for our calculations of sound correspondences, we would loose important information, as the tree part is also cognate in most instances and therefore rather interesting. But ignoring only the unalignable part of the first syllable in Mazhelong would also not be satisfying, since we would again have gaps for this word in the tree part in Mazhelong which do not result from sound change.
The only consistent solution to handle these cases is to split the words into their morphemes, and then to align all sets of homologous morphemes separately. This can also be done in the EDICTOR tool (but it requires more effort from the scholar and the algorithms). An example is shown above, where you can see how the tool breaks the linear order in the representation of the words as we find them in the languages, in order to cluster them into sets of homologous "word-parts".
 |
| Figure 3: Alignments of partial cognates in the Bai dialects |
But if we only look at the tree part of those alignments, namely the third cognate set from the left, with the ID 8, we can see a further complication, as the gaps introduced in some of the words look a little bit unsatisfying. The reason is that the j in Enqi and Tuolo may just as well be treated as a part of the initial of the syllable, and we could re-write it as dj in one segment instead of using two. In this way, we might capture the correspondence much more properly, as it is well known that those affricate initials in the other dialects (
[ts, tʂ, dʐ, dʑ]) often correspond to [dj].
We could thus rewrite the alignment as shown in the next figure, and simply decide that in this situation (and similar ones in our data), we
treat the d and the j as just one main sound (namely the initial of the syllables). |
| Figure 4: Revised alignment of "tree" in the sample |
Summary and conclusions
Before I start boring those of the readers of this blog who are not linguists, and not particularly interested in details of sound change or language change, let me just quickly summarize what I wanted to illustrate with these examples. I think that the reason why linguists never really formalized alignments as a tool of analysis is that there are so many ways to come up with possible alignments of words, which may all be reasonable for any given analysis. In light of this multitude of possibilities for analysis, not to speak of historical linguistics as a discipline that often prides itself by being based on hard manual labor that would be impossible to achieve by machines, I can in part understand why linguists were reluctant to use alignments more often in their research.
Judging from my discussions with colleagues, there are still many misunderstandings regarding the purpose and the power of alignment analyses in historical linguistics. Scholars often think that alignments directly reflect sound change. But how could they, given that we do not have any ancestral words in our sample? Alignments are a tool for analysis, and they can help to identify sound change processes or to reconstruct proto-forms in unattested ancestral languages; but they are by no means the true reflection of what happened and how things changed. The are the starting point, not the end point of the analysis. Furthermore, given that there are many different ways in which we can analyze how languages changed over time, there are also many different ways in which we can analyze language data with the help of alignments. Often, when comparing different alignment analyses for the same languages, there is no simple right and wrong, just a different emphasis on the initial analysis and its purpose.
As David wrote in an email to me:
"An alignment represents the historical events that have occurred. The alignment is thus a static representation of a dynamic set of processes. This is ultimately what causes all of the representational problems, because there is no necessary and sufficient way to achieve this."
This also nicely explains why alignments in biology as well, with respect to the goal of representing homology, "may be more art than science" (Morrison 2015), and I admit that I find it a bit comforting that biology has similar problems, when it comes to the question of how to interpret an alignment analysis. However, in contrast to linguists, who have never really given alignments a chance, biologists not only use alignments frequently, but also try to improve them.
If I am allowed to have an early New Year wish for the upcoming year, I hope that along with the tools that facilitate the labor of creating alignments for language data, we will also have a more vivid discussion about alignments, their shortcomings, and potential improvements in our field.
References
- Dixon, R. and A. Kroeber (1919) Linguistic families of California. University of California Press: Berkeley.
- List, J.-M. (2017) A web-based interactive tool for creating, inspecting, editing, and publishing etymological datasets. In: Proceedings of the 15th Conference of the European Chapter of the Association for Computational Linguistics. System Demonstrations, pp. 9-12.
- LingPy: A Python library for historical linguistics
- Morrison, D. (2015) Molecular homology and multiple-sequence alignment: an analysis of concepts and practice. Australian Systematic Botany 28: 46-62.
- Wang, W.-Y. (2006) Yǔyán, yǔyīn yǔ jìshù \hana 語言,語音與技術 [Language, phonology and technology]. Xiānggǎng Chéngshì Dàxué: Shànghǎi 上海.
Tuesday, December 12, 2017
Using consensus networks to understand poor roots
The standard way to root a tree is by outgroup rooting. Comprehensive outgroup sampling should minimize bias, such as ingroup-outgroup long-branch attraction (IO-LBA); but this may be impossible to avoid, when ingroup and all possible outgroups are very distant from each other. Using a recent all-Santalales up-to-7-gene dataset (Su et al. 2015) as an example, I will show how bootstrap consensus networks can illuminate topological issues associated with poorly supported roots.
These analyses should be obligatory whenever critical branches lack unambiguous support.
Some background: nothing to see, all is solved
The reason, why I had to re-analyse some of the data of Su et al., was that we dared to discuss alternatives to the accepted Loranthaceae (hereafter "loranth") root in our paper on loranth pollen (Grímsson, Grimm & Zetter 2017). Mainly for the artwork, I harvested gene banks for data on loranths in 2014 (re-checked in 2016, but no-one is studying this highly interesting group anymore). Earlier studies had used different taxon and gene sets, and produced cladograms (trees without branch lengths).
To assess the systematic value of loranth pollen, we needed to know how well resolved are intra-family relationships. Many deeper branches seen in the trees were not supported. What would be the competing alternatives? Unfortunately, one of our reviewers (#1), a veteran expert on plant parasites including loranths but not phylogenetics, was tremendously alienated by our tree, notably, the non-supported parts. He refused to understand the difference between a simple tree topology and branch support. Matrices producing ambiguous signals not-rarely lead to optimized trees that do not show the best-supported topological alternatives. Hence, for mapping our pollen, we did not use our tree, but the ML bootstrap consensus network (Fig. 1; which #1 refused even to look at). Fig. 2 shows the first plot focusing on the deepest, ambiguous relationships.
 |
| Fig. 1 The basis for our paper: a ML bootstrap consensus network, rooted under the commonly accepted assumption that the monotypic Nuytsia represents the first-diverging lineage. |
Reviewer #1 pointed to the recent paper of Su et al., which (according to his interpretation) resolved the intrafamily relationships. Since our tree was topologically somewhat different, and did not provide the same level of support, it had to be wrong. My argument was that the genetic data are inconclusive regarding critical branches in the loranth subtree. This is obvious from our bootstrap network (not including any outgroup, to avoid ingroup-outgroup branching artefacts), and also from the earlier molecular phylogenies, including Su et al.’s. Plus, to focus on the loranths we included the best-sampled and most-divergent plastid region, a non-coding region, which was not included in Su et al. because it is unalignable across the order.
A particularly "absurd" notion (according to #1) was that we suggested using pollen morphology to pre-define an alternative root (the best-fitting alternative according to Bayes factors; Grímsson, Kapli et al. 2017). All loranths share a unique, easy to distinguish, triangular pollen morph (Fig. 1) that can be traced back till the Eocene, except for one monotypic genus: Tupeia (antarctica) from New Zealand (ie. at the margin of their modern distribution). Tupeia’s pollen is spheroidal (Fig. 1), the basic form found all over the Santalales tree (Fig. 3). Fig. 4 shows alternative rooting scenarios using Su et al.'s loranth subtree.
 |
| Fig. 3 Schematic drawings of Santalales pollen, mapped on the preferred tree of Su et al. (cf. Grímsson, Zetter & Grimm, 2017, pp. 36–41) |
Why I lacked confidence in the Su et al. tree(s)
Su et al.’s study establishes relationships within the Santalales using a quite gappy up-to-7-gene data set (3 nuclear + 3 plastid + 1 mitochondrial genes) to revisit the placement of an originally poorly defined family with extremely long-branching members, the Balanophoraceae s.l. They found two main clades, one – the resurrected Mystropetalaceae – reconstructed as sister of the loranths (see scheme in Fig. 3). From what the authors show (but do not discuss), there are a lot of signal issues with their data:
- The standard nucleotide tree (partition scheme unclear) in the main text differs in many critical aspects (branching patterns, root-tip length ratios, support values) from the two in the supplement (one based on removing the 3rd codon positions, and one amino acid based).
- Many branches have high posterior-probabilities but low ML bootstrap values (BS), a pattern typical for mixed-genome oligogene data sets with intrinsic conflict.
- The first three branches within the loranth subtree, defining a ‘root parasitic grade’, have low to high posterior probabilities (PP = 0.65 / 0.99 / 0.52) and consistently low ML bootstrap supports (BS = <50 / 53 / <50).
- The sister clades have extremely long branches, and the putative sister clade, the Mystropetalaceae, is only covered for nuclear and mitochondrial data. All probabilistic inference methods treat missing data and gaps as “N”; ie. the terminal probability vector p equals (1,1,1,1) for p(A,C,G,T). In case the signal from the best-sampled regions is clear, missing data is no issue; if there is conflict then missing data can be a problem.
What’s behind the ambiguous bootstrap supports
For my head-to-head with the editor and reviewers (the editor had invited a #3 reviewer, an anonymous “expert on phylogeny”), I re-analysed Su et al.’s data subset on loranths plus sister groups. By reducing the outgroup sample to the sister clades only (Misodendraceae+Schoepfiaceae, Mystropetalaceae), I increased the BS support (100 / 62 / 60) for the root parasitic grade. This indicates either that Su et al.’s analysis was not comprehensive or that IO-LBA (ingroup-outgroup long-branch attraction) is prominent.
Large outgroup samples, all other Santalales + few non-Santalales in Su et al. vs. only loranths and sister clades in my re-analysis, may alleviate LBA to some degree (eg. Sanderson et al. 2000; Stefanović, Rice & Palmer 2004). Higher support with fewer outgroups than with many in the same dataset can be an alarm flag.
The ingroup-only bootstrap consensus network (Fig. 5) shows moderate, but unchallenged support for a split between root and aerial parasites, which agrees with Su et al.’s all-Santalales tree. The low support for the first and second branch in outgroup-rooted trees relates to an ambiguous signal between the root parasites — the signal from the combined data are ambiguous as to whether Atkinsonia (the second Australasian root parasite) or Gaiadendron (the South American root parasite) is the second diverging branch. Note that the position of our (pollen-based) alternative root candidate Tupeia is also unresolved (no split with a BS > 20; compare Figs 1 and 5), which could be expected for a potentially ancestral, relatively underived taxon — a taxon literally close to the family's root.
For most other deeper branches with high PP but low BS in Su et al.’s loranth subtree, the network shows two competing topological alternatives. There is thus an intrinsic signal conflict in Su et al.’s matrix, which explains the topological differences in earlier studies using different gene and taxon sets, preferring the one or other alternative in the final cladogram.
How to deconstruct a tree:
First step — single-gene trees and support networks
There are two main reasons for inferring single-gene trees and establishing single-gene support:
- It is the only way assess the level of incongruence in the gene samples.
- They inform about the amount of signal that each gene region contributes to the combined tree.
It is also clear that the nuclear and plastid data fit aspect-wise, eg. both support or at least don’t reject potential clades, but not entirely. Particularly, the placements of the sister groups of the loranths within the loranth tree are unstable, ie. the position of the outgroup-inferred root.
Last, the single-gene trees and bootstrap networks illustrate a severe taxon sampling bias. The third nuclear marker, the RBS2 gene, is only covered for five ingroup taxa including Nuytsia but no other root parasite, and it fails to resolve the two aerial tribes, which are otherwise genetically distinct. Analysis of this small subset illustrates nicely the perils of taxon sampling (Fig. 7). The third plastid gene, the accD gene (7 ingroup representatives), supports Nuytsia as the first diverging lineage, but rejects the root parasite grade with equally high support (BS = 87)
The only mitochondrial gene included in the data set (matR) rejects the root-parasitic grade, in addition to the matK-only based Elytrantheae-Lorantheae clade seen in Su et al.’s tree, with nearly unambiguous support. However, it is in general low-divergent, and it may be that the authors relied on fragmentary (potentially pseudogene) data for some taxa. There are huge indels in the gene not found in any other angiosperm, and the two sequenced Loranthus are conspicuously distinct to the other loranths (Fig. 8).
One of the arguments of reviewer #1 was that Su et al. obtained a better resolved phylogeny because they sampled these extra gene regions. In fact, they obtained a better supported phylogeny because they added diffuse-signal or under-sampled gene regions. Hence, they enforced the dominant matK-preferred topology (high PP, low BS), while weaking locally conflicting signal from the nuclear 18S and 25S rDNA.
The only constant is that, no matter which gene region, Nuytsia is always the most distinct taxon of all species close to the root node. With respect to the very distant sister clades, IO-LBA may be inevitable. The BS consensus networks favor a closer relationship of the root parasites, and there is relatively strong signal for a root parasitic clade in the matK (Fig. 5), so that the biased outgroup-root then becomes a proximal (‘basal’) grade (Fig. 6, left).
Data effects of the LBA-missing sort also challenge the placement of the Mystropetalaceae as sister to the loranths. The plastid data establish that Misodendraceae and Schoepfiaceae (M+S) are distinct from the loranths. They provide unambiguous support for a loranth versus M+S split. Having no data on any plastid gene, the extremely long-branched Mystropetalaceae fall in line. Being very distinct from both M+S and loranths in the nuclear (Fig. 5) and mitochondrial (Fig. 8) genes, the tree places them next to the loranths, because the latter's root node and many of its members are much more distant from the all-ancestor than those of the M+S clade (compare with Su et al.'s trees; keep in mind that missing data may increase branch-lengths).
Second step—remove a taxon, or two, or a gene region
If the hypothesis of a root-parasitic grade holds, then one should be able to remove the first diverging taxon (Nuytsia in this example) from the dataset, and still have the position of the second (Atkinsonia) and third diverging (Gaiadendron) remain the same. Furthermore, the support for the corresponding branches should at best increase, and at least not decrease. Atkinsonia is relatively distinct, but Gaiadendron is not (as reflected by their pollen morphology, Fig. 2).
In the case of Su et al.’s data, removal of Nuytsia collapses the root-proximal part of the ingroup tree (Fig. 9). ML-tree inferences select a sub-optimal topology, because the signal weakens for the splits {Atkinsonia + outgroups | all other Loranthaceae} and {root parasites + outgroups | aerial parasites}. The competing alternative (BS = 23) is a first-diverging Atkinsonia-Gaiadendron clade. When both Nuytsia and Atkinsonia are removed, Gaiadendron is placed within the aerial parasites, and the Elytrantheae, a low-diverged Australasian tribe, are favored as first-diverging loranth clade (BS increases from 15 to 39; any alternative BS is < 20; see also their placements in Fig. 5).
Equally disastrous for the support of a root parasitic grade is the elimination of matK from the combined data set. This is a major advantage of BS or support consensus networks: one can estimate whether taxon or gene sampling increases or decreases support for competing topologies; and not only the combined trees’ topologies.
Support consensus networks should be obligatory when studying non-trivial or suboptimal data sets
When it comes to extremely long-branching roots and sister groups, one should generally be careful with combined-data roots defined by outgroups (see also our discussion of the outgroup-inferred Osmundaceae root in Bomfleur, Grimm & McLoughlin 2015). But when the crucial branches in addition lack consistent support, one should actually be alarmed.
In the case of gappy oligogene (as used by Su et al.) or multigene data, one should always test the primary signal in the combined gene regions. Fast and efficient ML implementations make it very easy to infer and compare single-gene trees, and thus establish differential branch support patterns, which can be visualized and investigated using support consensus networks. As biologists working with complex systems and, likely, non-trivial evolutionary pathways, we should not be afraid of conflicting splits patterns, but deal with them!
It can also be useful to plot (tabulate) competing support values from different analysis. RAxML has an option that allows testing the direct correlation between two bootstrap (or Bayesian) tree samples, without being restricted to any particular tree. The Phangorn library for R includes functions to map trees into networks, and transfer support from networks to trees. In my experience, many nuclear and plastid incongruences are:
- tree-based, meaning the trees are different, but the supports for topological alternatives are not conflicting;
- resolution-based, meaning the nuclear data illuminate different parts of the evolutionary history compared to the plastid data;
- rogue-based, meaning there are only a few taxa with strongly conflicting signals in the tree.
Uploading a matrix and the optimized tree to (eg.) TreeBase is just the first step (although many TreeBase submissions include only naked trees). In the case of ambiguous branch support (BS < 80 or 85, depending on the proportion of nuclear vs. mitochondrial and plastid gene regions; PP < 1.00), much more important would be to document the bootstrap replicate and Bayesian tree samples.
Furthermore, reviewers and editors should not encourage the publishing of trees where low support is masked by cut-offs (eg. BS < 50, PP < 0.95). Which is, unfortunately, the editorial policy of the journal that published the Su et al. study, and the reason for the scarcity of studies showing support consensus or other networks that can illuminate signal issues (but see eg. Denk & Grimm 2010, Wanntorp et al. 2014, and Khanum et al. 2016, published in Taxon). The latter a common feature in plant molecular data sets, independent of the taxonomic hierarchical level.
Further reading, and data
The full details of my re-analysis of Su et al.’s data subset, including loranths and sister clades, can be found in File S6 [PDF] to Grímsson, Grimm & Zetter (2017), included in the Online Supporting Archive (9.5 MB; journal hompage/mirror]. The relevant data and analysis files can be found in subfolder "Su_el_al." of the archive.
References
Bomfleur B, Grimm GW, McLoughlin S (2015) Osmunda pulchella sp. nov. from the Jurassic of Sweden — reconciling molecular and fossil evidence in the phylogeny of modern royal ferns (Osmundaceae). BMC Evolutionary Biology 15: 126. http://dx.doi.org/10.1186/s12862-015-0400-7
Denk T, Grimm GW. 2010. The oaks of western Eurasia: traditional classifications and evidence from two nuclear markers. Taxon 59:351–366. — Includes neighbour-nets based on inter-individual and uncorrected p-distances, with ML branch support mapped and tabulated.
Grímsson F, Grimm GW, Zetter R (2017) Evolution of pollen morphology in Loranthaceae. Grana DOI:10.1080/00173134.2016.1261939. http://dx.doi.org/10.1080/00173134.2016.1261939
Grímsson F, Kapli P, Hofmann C-C, Zetter R, Grimm GW (2017) Eocene Loranthaceae pollen pushes back divergence ages for major splits in the family. PeerJ 5: e3373 [e-pub]. https://peerj.com/articles/3373/ Don't miss reading the peer review documentation, which includes some interesting discussion. (If you agree that the peer review should be transparent in general, then you can sign up here for fighting the windmills.)
Khanum R, Surveswaran S, Meve U, Liede-Schumann S. 2016. Cynanchum (Apocynaceae: Asclepiadoideae): A pantropical Asclepiadoid genus revisited. Taxon 65:467–486. — Includes trees, median-joining and bootstrap consensus networks [I really enjoyed working on this, but retreated from co-authorship to protest against Taxon's editorial policies.]
Sanderson MJ, Wojciechowski MF, Hu J-M, Sher Khan T, Brady SG (2000) Error, bias, and long-branch attraction in data for two chloroplast photosystem genes in seed plants. Molecular Biology and Evolution 17: 782-797.
Stefanović S, Rice DW, Palmer JD (2004) Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evolutionary Biology 4: 35. http://biomedcentral.com/1471-2148/4/35
Su H-J, Hu J-M, Anderson FE, Der JP, Nickrent DL (2015) Phylogenetic relationships of Santalales with insights into the origins of holoparasitic Balanophoraceae. Taxon 64: 491-506.
Vidal-Russell R, Nickrent DL (2008) The first mistletoes: origins of aerial parasitism in Santalales. Molecular Phylogenetics and Evolution 47: 523-537.
Wanntorp L, Grudinski M, Forster PI, Muellner-Riehl AN, Grimm GW. 2014. Wax plants (Hoya, Apocynaceae) evolution: epiphytism drives successful radiation. Taxon 63:89–102. — Includes trees and a "splits rose" [The editors forced us do drop some quite nice bootstrap consensus and median-joining networks; see also this post]
Tuesday, December 5, 2017
The Synoptic Gospels problem: preparing a phylogenetic approach
This is the second part of my series on phylogenetics and a specific case of textual criticism, the Biblical one. The first part appeared as Another test case for phylogenetics and textual criticism: the Bible, and covered the background to the textual problem — that post should be read first. Here, I provide a preliminary genealogical analysis of some specific data related to the problem.
The synoptic gospels and phylogenetics: how to code data?
Just like in the cases of general stemmatics and historical linguistics, our immediate problem for a phylogenetic approach to Biblical criticism is one of data. Upon investigation, the field proves itself desperately in need of an open access mentality — a great deal of work would be needed to turn the few aggregated data I could find into datasets that could feed the most basic analysis tools.
No open dataset proved either adequate or correct enough. They are mostly quotations or subjective developments of the scientific sources, available only in printed editions and in software for Biblical studies, sometimes at exorbitant prices, and frequently with licenses that explicitly prohibit extracting and reusing the data. This forced me to postpone an analysis of families of manuscripts, as unfortunately there is no complete free edition of the Novum Testamentum Graece (the reference work in the field, usually referred to as Nestle-Aland after its main editors).
However, I could explore the problem of the synoptic gospels in a way and with a dataset closer to the ones of the 19th century analyses, by sitting with a printed Bible and compiling my own synopsis of episodes. My work in this field ends with this second post, but it seems like a good approach to the development of a phylogenetic investigation, to start by reproducing the old analyses with new tools.
After some bibliographic review and inspection of the solutions presented to the problem, my understanding is that there would be three fundamental ways of coding for features of these texts.
(i)
The first and simplest is to compile a list of episodes, themes, and topics found in each gospel (a proper “synopsis”), without considering semantic differences or relative positions, coding for a truth table indicating whether each “event” (i.e. “character”) is found. For example, the imprisonment of John the Baptist is mentioned in the three synoptic gospels (Matthew 4,12; Mark 1,14; Luke 3,18-20) and would be coded as “present” in all of them, even though in Luke the relative order is different (it is narrated before the baptism of Jesus, in a flashforward). On the other hand, the priests conspiring against Jesus is only narrated in two gospels (Mark 11,18; Luke 19,47-48), and the “character” of the meek inheriting the Earth is only found in one of them (Matthew 5,5), as shown in the table below.
Character/Event
| Matthew | Mark | Luke |
Imprisonment of John
| ✓ | ✓ | ✓ |
Priests conspiring
| ✓ | ✓ | |
Meek inheritance
| ✓ |
This kind of census approach is what most descriptive statistics on the synoptic relationship consider when demonstrating how much there is in common among the gospels, including the graph reproduced back in the first part of this post. As in the case of the statistics of genetic material shared between species, like humans and other apes, caution is needed to understand what is actually meant — the percentages usually reported refer to episode coincidence (in a loose analogy, like the presence of a protein), not text coincidence (like the sequences of genetic bases). This is the reason why these analyses should equally consider “episode homology” and “episode analogy” — one must remember that all gospels as we have them evolved from initial versions, and to be missing an episode favored by the public or the clergy, which denounced other gospels now lost as “uninspired”, could have resulted in pressure to incorporate such episode.
(ii)
A deeper level of coding would be to map the text of episodes and events into “semantic” characters, ignoring textual differences (like synonyms) but coding for differences in intended meaning. For example, the event of Jesus being tested in the wilderness, while narrated in all three gospels (Matthew 4,1-2; Mark 1,12-13; Luke 4,1-2), is really only equivalent in Matthew and Luke, where he is tempted by "the Devil", while in Mark he is tempted by "Satan", which is a figure closer to the Hebrew meaning of "enemy, adversary; accuser". Likewise, while Matthew and Luke both narrate Jesus’ most famous sermon, they are semantically different: the setting is a mountain in the first and a plain in the second.
Character/Event
| Matthew | Mark | Luke |
Temptation
| by the Devil | by Satan | by the Devil |
Sermon
| mountain | plain |
This kind of mapping is harder, due to the expertise required to subjectively distinguish meaning, as in the case of the mountain / plain, which scholars in Biblical hermeneutics seem to agree to be more than merely a change of setting for narration. The difficulty is aggravated by the eventual need to quantify the semantic shifts (how far is "the Devil" from "Satan (the adversary)", especially when the episode is missing from the non-synoptic gospel of John?). These three states ("null", "Devil", and "Satan") should not be considered equally different, especially when the texts of the three synoptic gospels are clearly related. Luckily, while not necessarily in a systematic way for phylogenetic purposes, this kind of coding has already been conducted by many Biblical scholars, and we might thus appropriate it in the future.
(iii)
The third way of coding, partly solving the difficulties of the second solution, would be to compare the Greek text for each event, using some distance metric. For strings, there is the common Levenshtein distance, or, in a blatant self-promotion, my own sequence similarity algorithm. For linguistic texts, there are dozens of possible Natural Language Processing solutions, but usually with no model for Koine Greek (apart from purely statistical ones that can overfit, because in general they are actually trained on the text of the gospels, in the first place).
| Character/Event | Matthew | Mark | Luke |
Prologue
| Βίβλος γενέσεως Ἰησοῦ... (1,1) | Ἀρχὴ τοῦ εὐαγγελίου Ἰησοῦ... (1,1) | Ἐπειδήπερ πολλοὶ ἐπεχείρησαν... (1,1-4) |
Birth of Jesus
| Τοῦ δὲ Ἰησοῦ χριστοῦ ἡ γένεσις... (1,18-25) | Ἐγένετο δὲ ἐν ταῖς ἡμέραις ἐκείναις... (2,1-7) | |
Healing of possessed
| καὶ εὐθὺς ἦν ἐν τῇ συναγωγῇ αὐτῶν ἄνθρωπος ἐν πνεύματι... (1,23-28) | καὶ ἐν τῇ συναγωγῇ ἦν ἄνθρωπος ἔχων... (4,33-37) | |
Parable of tares
|
Ἄλλην παραβολὴν παρέθηκεν αὐτοῖς λέγων... (13,24-30)
|
By comparing all distance pairs for all characters, we could build a matrix of pairwise distances, similarly to what David frequently does in the EDA analyses posted to this blog. Considering that most synoptic lists have already mapped each event to their texts (sometimes in discontinuous blocks), with a copy of the reconstructed Greek original, from Holmes (2010) in the table immediately above, it should not be too hard to perform such a study.
For the purpose of this post, I decided to proceed with the first of these three possible solutions, listing whether an event is found in each Gospel or not, ignoring semantic and textual differences. I modified the synopsis by Garmus (1982), itself apparently modified from some Nestle-Aland edition. This produced a final list of 364 characters and their presence in each of the four gospels — I decided to include the non-synoptic John to test where the analyses would place it.
As expected, the data are to a large extent arbitrary and subjective. Garmus has obvious limitations in the way of dealing with events narrated out of the expected chronological sequence (i.e., flashbacks and flashforwards, as in the case of the beheading of John in relation to his actions), as well as with theological excursuses. None of these limits, however, seem to impact the general shape of a network or tree generated from these data, at most strengthening more feeble signals.
 |
| Splits tree, modified from the one generated by Huson & Bryant (2010) |
As also expected, the graph supports what is by now a general consensus. Mark is likely to be the gospel closest to a hypothetical root (in this case, nearest to the mid-point). John is the most distinct of the four gospels, being closer to Mark than to the Matthew-Luke group (due to the “core” events narrated and the fewer innovations in Mark). Considering edge lengths, Luke seems to be the most innovative taxon of the synoptic gospel neighborhood / group. Such a network could never demonstrate the existence of "Q" (see the first post) as a stand-alone and actual document, but this tentative analysis does support the hypothesis that Matthew and Luke share a common development, overall supporting Marcan priority.
While probably obvious, it is important to remember that phylogenetic methods are tools that imply the existence of users — it should be an additional instrument for investigation, possibly promoting the collaboration of serious Biblical critics and experts in phylogenetic methods. Let’s consider two examples of the need for such expertise.
First, there is much historical, textual, and theological evidence supporting a hypothesis that the gospel of Mark originally ended with what is now Mark 16,8, with the twelve following verses as later additions (something common to many Greek texts, including the Odyssey). If these supposed additions, only known to whoever delves into Biblical scholarship, are marked as missing in our data, as we should at least test, the distance between Mark and all other gospels, including the unrelated Gospel of John and especially in the edge length between Luke and Mark, increases considerably for such an apparently minor change.
Second, if conducting the third and especially the second type of coding that I described above, a researcher should have at least a basic knowledge of the language they are dealing with. Adapting the explanation of Smith (2017), Matthew and Mark might seem to use the same vocabulary for the “parable of the harvest” when read in English translation, but there is a concealed change of meaning (whose theological importance and implication I'm not debating here), as the single English word “seed” tends to be used in translation of two different Greek words: in Matthew, “sperma” (the kernels of grain, in a more agricultural sense) and, in Mark, “sporos” (which carries a connotation of generative matter to be released).
Conclusion
My dataset is available in preliminary state (for example, labels are in Portuguese) here.
In conclusion, phylogenetics still has much to offer to the field of textual criticism, and this should include Biblical criticism, especially if we are able to support analyses of textual development based on trees / networks of manuscripts. I hope this pair of blog posts will motivate Biblical scholars to collaborate. If so, please write to me.
References
Garmus, Ludovico (ed.) (1982) Bíblia sagrada. Petrópolis: Editora Vozes. [reprint 2001]
Goodacre, Mark (2001) The Synoptic Problem: a Way Through the Maze. New York: T & T Clark International. (available on Archive.org)
Holmes, Michael W. (ed.) (2010) SBL Greek New Testament. Atlanta, GA: Society of Biblical Literature.
Huson, Daniel H.; Bryant David (2006) Application of Phylogenetic Networks in Evolutionary Studies, Mol. Biol. Evol., 23(2):254-267. [SplitsTree.org]
Smith, Mahlon H (2017) A Synoptic Gospels Primer. http://virtualreligion.net/primer/
Tuesday, November 28, 2017
“Man gave names to all those animals”: goats and sheep
This is a joint post by Guido Grimm, Johann-Mattis List, and Cormac Anderson.
This is the second of a pair of posts dealing with the names of domesticated animals. In the first part, we looked at the peculiar differences in the names we use for cats and dogs, two of humanity’s most beloved domesticated predators. In this, the second part (and with some help from Cormac Anderson, a fellow linguist from the Max Planck Institute for the Science of Human History), we’ll look at two widely cultivated and early-domesticated herbivores: goats and sheep.
Similar origins, but not the same
Both goats and sheep are domesticated animals that have an explicitly economic use; and, in both cases, genetic and archaeological evidence points to the Near East as the place of domestication (Naderi et al. 2007). The main difference between the two is the natural distribution of goats (providing nourishment and leather) and sheep (providing the same plus wool). This distribution is also reflected in the phonetic (dis)similarities of the terms used in our sample of languages (Figures 1 and 2).
Capra aegagrus, the species from which the domestic goat derives, is native to the Fertile Crescent and Iran. Other species of the genus, similar to the goat in appearance, are restricted to fairly inaccessible areas of the mountains of western Eurasia (see Figure 3, taken from Driscoll et al. 2009). On the other hand, Ovis aries, the sheep and its non-domesticated sister species, are found in hilly and mountainous areas throughout the temperate and boreal zone of the Northern Hemisphere. Whenever humans migrated into mountainous areas, there was the likelihood of finding a beast that:
Had wool on his back and hooves on his feet,Goats
Eating grass on a mountainside so steep
[Bob Dylan: Man Gave Names to all those animals].
Goats were actively propagated by humans into every corner of the world, because they can thrive even in quite inhospitable areas. Reflecting this, differences in the terms for "goat" generally follow the main subgroups of the Indo-European language family (Figure 1), in contrast to "cat", "dog", and "sheep". From the language data, it seems that for the most part each major language expansion, as reflected in the subgroups of Indo-European languages, brought its own term for "goat", and that it was rarely modified too much or borrowed from other speech communities.
There is one exception to this, however. The terms in the Italic and Celtic languages look as though they are related, coming from the same Proto-Indo-European root, *kapr-, although the initial /g/ in the Celtic languages is not regular. In Irish and Scottish Gaelic, the words for "sheep" also come from the same root. In other cases, roots that are attested in one or other language have more restricted meanings in some other language; for example, the Indo-Iranic words for goat are cognate with the English buck, used to designate a male goat (or sometimes the male of other hooved animals, such as deer).
The German word Ziege sticks out from the Germanic form gait- (but note the Austro-Bavarian Goaß, and the alternative term Geiß, particularly in southern German dialects). The origin of the German term is not (yet) known, but it is clear that it was already present in the Old High German period (8th century CE), although it was not until Luther's translation of the Bible, in which he used the word, that the word became the norm and successively replaced the older forms in other varieties of Germany (Pfeifer 1993: s. v. "Ziege").
 |
| Figure 1: Phonetic comparison of words for "goat" |
Sheep
The terms for sheep, however, are often phonetically very different even in related languages. The overall pattern seems to be more similar to that of the words for dog – the animal used to herd sheep and protect them from wolves. An interesting parallel is the phonetic similarity between the Danish and Swedish forms får (a word not known in other Germanic languages) and the Indic languages. This similarity is a pure coincidence, as the Scandinavian forms go back to a form fahaz- (Kroonen 2013: 122), which can be further related to Latin pecus "cattle" (ibd.) and is reflected in Italian
[pɛːkora]
in our sample.This example clearly shows the limitations of pure phonetic comparisons when searching for historical signal in linguistics. Latin c (pronounced as
[k])
is usually reflected as an h in Germanic languages,
reflecting a frequent and regular sound change. The sound [h]
itself can be easily lost, and the [z]
became a [r]
in many Scandinavian words. The fact that both Italian and Danish plus
Swedish have cognate terms for "sheep", however, does not
mean that their common ancestors used the same term. It is much more
likely that speakers in both communities came up with similar ways to
name their most important herded animals. It is possible, for
example, that this term generically meant "livestock", and
that the sheep was the most prototypical representative at a certain
time in both ancestral societies.Furthermore, we see substantial phonetic variation in the Romance languages surrounding the Mediterranean, where both sheep and goats have probably been cultivated since the dawn of human civilization. Each language uses a different word for sheep, with only the Western Romance languages being visibly similar to ovis, their ancestral word in Latin, while Italian and French show new terms.
 |
| Figure 2: Phonetic comparison of words for "sheep" |
More interesting aspects
The wild sheep, found in hilly and mountainous areas across western Eurasia, was probably hunted for its wool long before mouflons (a subspecies of the wild sheep) were domesticated and kept as livestock. The word for "sheep" in Indo-European, which we can safely reconstruct, was h₂ owis, possibly pronounced as
[xovis],
and still reflected in Spanish, Portuguese, Romanian, Russian,
Polish. It survives in many more languages as a specific term with a
different meaning, addressing the milk-bearing / birthing female sheep. These include
English ewe, Faroean ær (which comes
in more than a dozen
combinations; Faroes literally means: “sheep islands”),
French brebis (important
to known when you want sheep-milk based cheese), German Aue (extremely rare nowadays, having been
replaced by Mutterschaf "mother-sheep"). In
other languages it has been lost completely.What is interesting in this context is that while the phonetic similarity of the terms for "sheep" resembles the pattern we observe for "dog", the history of the words is quite different. While the words for "dog" just continued in different language lineages, and thus developed independently in different groups without being replaced by other terms, the words for "sheep" show much more frequent replacement patterns. This also contrasts with the terms for "goat", which are all of much more recent origin in the different subgroups of Indo-European, and have remained rather similar after they were first introduced.
The reasons for these different patterns of animal terms are manifold, and a single explanation may never capture them all. One general clue with some explanatory power, however, may be how and by whom the animals were used. Humans, in particular nomadic societies, rely on goats to colonize or survive in unfortunate environments, even into historic times. For instance, goats were introduced to South Africa by European settlers to effectively eat up the thicket growing in the interior of the Eastern Cape Province. Once the thicket was gone, the fields were then used for herding cattle and sheep.
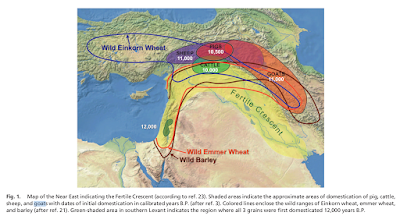 |
| Figure 3: Map from Driscoll et al. (2009) |
There are other interesting aspects of the plot.
For example, as mentioned before, in Chinese the goat refers to the "mountain sheep/goat" and the "sheep/goat" is the "soft sheep". While it is straightforward to assume that yáng, the term for "sheep/goat", originally only denoted one of the two organisms, either the sheep or the goat, it is difficult to say which came first. The term yáng itself is very old, as can also be seen from the Chinese character used, which serves as one of the base radicals of the writing system, depicting an animal with horns: 羊. The sheep seems to have arrived in China rather early (Dodson et al. 2014), predating the invention of writing, while the arrival of the goat was also rather ancient (Wei et al. 2014) (and might also have happened more than once). Whether sheep arrived before goats in China, or vice versa, could probably be tested by haplotyping feral and locally bred populations while recording the local names and establishing the similarity of words for goat and sheep.
While the similar names for goat and sheep may be surprising at first sight (given that the animals do not look all that similar), the similarity is reflected in quite a few of the world's languages, as can be seen from the Database of Cross-Linguistic Colexifications (List et al. 2014) where both terms form a cluster.
Source Code and Data
We have uploaded source code and data to Zenodo, where you can download them and carry out the tests yourself (DOI: 10.5281/zenodo.1066534). Great thanks goes to Gerhard Jäger (Eberhard-Karls University Tübingen), who provided us with the pairwise language distances computed for his 2015 paper on "Support for linguistic macro-families from weighted sequence alignment" (DOI: 10.1073/pnas.1500331112).
Final remark
As in the case of cats and dogs, we have reported here merely preliminary impressions, through which we hope to encourage potential readers to delve into the puzzling world of naming those animals that were instrumental for the development of human societies. In case you know more about these topics than we have reported here, please get in touch with us, we will be glad to learn more.
References
- Dodson, J., E. Dodson, R. Banati, X. Li, P. Atahan, S. Hu, R. Middleton, X. Zhou, and S. Nan (2014) Oldest directly dated remains of sheep in China. Sci Rep 4: 7170.
- Driscoll, C., D. Macdonald, and S. O’Brien (2009) From wild animals to domestic pets, an evolutionary view of domestication. Proceedings of the National Academy of Sciences 106 Suppl 1: 9971-9978.
- Jäger, G. (2015) Support for linguistic macrofamilies from weighted alignment. Proceedings of the National Academy of Sciences 112.41: 12752–12757.
- Kroonen, G. (2013) Etymological dictionary of Proto-Germanic. Brill: Leiden and Boston.
- List, J.-M., T. Mayer, A. Terhalle, and M. Urban (eds) (2014) CLICS: Database of Cross-Linguistic Colexifications. Forschungszentrum Deutscher Sprachatlas: Marburg.
- Naderi, S., H. Rezaei, P. Taberlet, S. Zundel, S. Rafat, H. Naghash, et al. (2007) Large-scale mitochondrial DNA analysis of the domestic goat reveals six haplogroups with high diversity. PLoS One 2.10. e1012.
- Pfeifer, W. (1993) Etymologisches Wörterbuch des Deutschen. Akademie: Berlin.
- Wei, C., J. Lu, L. Xu, G. Liu, Z. Wang, F. Zhao, L. Zhang, X. Han, L. Du, and C. Liu (2014) Genetic structure of Chinese indigenous goats and the special geographical structure in the Southwest China as a geographic barrier driving the fragmentation of a large population. PLoS One 9.4: e94435.
Tuesday, November 21, 2017
Another test case for phylogenetics and textual criticism: the Bible
This is a two-part blog post. Here, I will introduce a particular stemmatological problem, along with the studies of it to date; and in a subsequent post I will discuss possible phylogenetic analyses that might be applied (see The Synoptic Gospels problem: preparing a phylogenetic approach).
Introduction
This year marks the celebration of 500 years since Martin Luther famously proposed his 95 religious theses, thus presaging the Protestant Reformation of the Western Christian Church. In line with this, it is worth discussing a subfield of textual criticism and stemmatics deeply influenced by the Reformation: Biblical criticism. While the importance of written texts to Christianity begins at least in the 2nd century, the theological doctrine of the sola fide (“by scripture alone”, regarding the infallible and final authority in all matters), along with translation work and individual study of the Bible, paved the way, sometimes unwillingly, to scientific approaches of Biblical criticism equivalent to those of secular literature.
The seminal figure in textual criticism of the New Testament was Hermann Reimarus (1694-1768), apparently the first to apply the methodology of literary texts to religious ones. As in the case of literary criticism, it is hardly a coincidence that Biblical criticism developed in the same cultural framework that would support and promote the idea of biological evolution and the tools for establishing genealogical trees and networks. This is especially so when considering the secularization of that society, in which proving the human origin and transmission of sacred texts was deemed an important act of civic freedom. Along with this was the parallel radicalization of some religious positions, such as denouncement as heresy of scientific studies of religious texts (nowadays objected to by most Christian doctrines that stated the imperative of serious research on the sacred texts).
A concrete problem: the synoptic gospels
The most important problem in the textual criticism of the New Testament is the “synoptic gospels" one, involving the three Gospels of Mark, Matthew, and Luke. These gospels have strikingly similar narratives that relate many of the same stories, with similar or identical wording. Like the other canonical gospel, John, these texts were composed around the last quarter of the first century by literate Greek-speaking Christians, only becoming canonical at least a century after their composition.
The synoptic gospels differ from similar sources, such as the non-canonical Gospel of Thomas, in being biographies with a clear religious motivation, and not just a collection of sayings. When compared to the Gospel of John, the three synoptic gospels are distinct in apparently being written by and for a Jewish community that was not on the verge of breaking from the Jewish synagogue, also favoring short and simple sentences.
However, the most important proof of their genealogical relationship is the text itself. The table below shows the reconstructed Greek original of each gospel for the episode of Jesus’ recruitment of a tax collector (an episode missing from the non-synoptic Gospel of John). The text in blue is the material shared by any two of the gospels, and the text in red is common to all three of them. [This is adapted from Smith (2017); on Wikipedia there is a further example, referring to the episode of the cleansing of a leper, see https://en.wikipedia.org/wiki/Synoptic_Gospels#Example.]
|
Matthew 9,9
|
Mark 2,13-14
|
Luke 5, 27-28
|
| Καὶ παράγων ὁ Ἰησοῦς ἐκεῖθεν εἶδεν ἄνθρωπον καθήμενον ἐπὶ τὸ τελώνιον, Μαθθαῖον λεγόμενον, καὶ λέγει αὐτῷ· Ἀκολούθει μοι. καὶ ἀναστὰς ἠκολούθ ησεν αὐτῷ. | Καὶ ἐξῆλθεν πάλιν παρὰ τὴν θάλασσαν· καὶ πᾶς ὁ ὄχλος ἤρχετο πρὸς αὐτόν, καὶ ἐδίδασκεν αὐτούς. καὶ παράγων εἶδεν Λευὶν τὸν τοῦ Ἁλφαίου καθήμενον ἐπὶ τὸ τελώνιον, καὶ λέγει αὐτῷ· Ἀκολούθει μοι. καὶ ἀναστὰς ἠκολούθ ησεν αὐτῷ. | Καὶ μετὰ ταῦτα ἐξῆλθεν καὶ ἐθεάσατο τελώνην ὀνόματι Λευὶν καθήμενον ἐπὶ τὸ τελώνιον, καὶ εἶπεν αὐτῷ· Ἀκολούθει μοι. καὶ καταλιπὼν πάντα ἀναστὰς ἠκολούθ ει αὐτῷ. |
The relationships between the gospels, such as the so-called “triple tradition”, is summarized by the graph below, from the Wikipedia article on the synoptic gospels. Mark, the shortest text, has almost no unique material (only 3%, in part superfluous adjectives and Aramaic translations) and is almost entirely (94%) reproduced in Luke. Matthew and Luke have their share of unique material (20% and 35%, respectively), which suggests independence, except for a "double tradition" of common material of about a quarter of the contents of each one, including notable passages such as the “Sermon of the Mount”. The parallelisms of these two gospels are found not only in their contents, but also in their arrangement, with most episodes described in the same order and, in case of displacements, with blocks of episodes moved together while preserving their internal order.
Previous studies
Such similarities were already noted in the first centuries of Christianity. This raises typical genealogical questions regarding topics such as priority (which gospel was written first) and dependence (which gospel was used as a source).
As for the first question, due to textual and theological evidence, a well-established majority of commentators favors the hypothesis of Marcan priority — that is, that the gospel of Mark is the oldest, and both Matthew and Luke used it as a source. As for the second question, a major point of dispute is the double tradition of Matthew and Luke, which can only be properly explained in terms either of descent or of a common ancestor. The two leading hypotheses are the one of a lost gospel (referred as “Q”, after the German Quelle [“source”]), and the one by Austin Farrer, according to whom Matthew used Mark as its source and Luke then used both of them. But these are not the only hypotheses that have been proposed, as shown in the next set of diagrams (also from the Wikipedia article above).
|
|
||||
|
|
The first fully developed theory was actually proposed by Augustine of Hippo back in the 5th century, which is essentially the one by Farrer, but with Matthew in place of Mark (i.e., supporting a Matthean priority). Given Augustine’s authority as a “Father of the Church”, his view was not disputed until the late 18th century, when Johann Jakob Griesbach published a synopsis of the three gospels and developed a new hypothesis, swapping Mark and Luke in the dominant explanation. Griesbach’s scientific approach led to the first application to Biblical problems of textual criticism, then in development in the German towns of Jena and Leipzig where he lived.
In 1838, Christian Weisse proposed the “Q” Hypothesis, mentioned above, asserting that Matthew and Luke were produced independently, both using Mark plus a lost source. This source was described as a lost collection of sayings of Jesus, along with feeble indirect evidence of its existence. This hypothesis was further developed by Burnett Streeter in 1924, with the proposal of “proto-versions” of both Mark and Luke — the wording of the canonical versions we have today would then be the product of later revisions, influenced by all of the texts.
During the past fifty years, due to advances in textual criticism and new manuscript analyses, the independence of Luke in relation to Matthew has been questioned, with diminishing support for the Q Hypothesis. A now leading position holds for Farrer’s hypothesis, along with alternative trees such as the one by the Jerusalem School, according to which a lost Greek anthology “A” (postulated as the translation of a collection of sayings either in Hebrew or in Aramaic) was directly or indirectly used by all gospels, including John.
Phylogenetics
Considering the analogies between literary and genetic texts that we have already discussed on this blog, it is clear that this topic should be an interesting anecdote to share around phylogenetic water-coolers. The four texts can be divided into two “families” of gospels, the synoptic (taxa: Matthew, Mark, and Luke) and non-synoptic (taxon: John). Their similarities suggest a distant common ancestor, probably oral traditions, as reported by Christian writers of the first and second centuries such as Papias.
The relationship between the taxa of the first family, however, is far from clear, as their relative dates cannot be determined with confidence. We might be faced with processes that, by analogy with biology, can be explained as gene pool recombinations and horizontal gene transfers – even though the most likely explanation is the one of direct descent, possibly from unknown taxa.
In literary terms, we must also consider features such as Matthew clearly being written by someone highly familiar with aspects of Jewish law, possibly asserting the Jewish component of the preaching while perceiving a universal tendency for the new faith. We must also consider the fact that Mark provides no ancestral lineage for Jesus, while Matthew traces him from a line of kings and Luke from a line of commoners — clearly stating the theological point of view of each gospel. Other aspects are worth consideration, such as the idea that what we today identify as the Gospel of Luke is likely to have been the first part of a once single document that included what is now the book of the “Acts of the Apostles”.
While I must admit that my research has been limited to some googling of keywords, it is curious that a topic that has attracted so much attention for millennia, from serious academic scholarship to conspiracy theories, and from impressionistic reviews to advanced statistical modeling, does not seem to have been covered by phylogenetic analyses, so far. Given the range of data and literature, it should actually look like a prime candidate for such application, even from an outsider point of view. This viewpoint is in fact discussed in a review by Christian P. Robert of a book called The Synoptic Problem and Statistics by Andry Abakuks:
The book by Abakuks goes […] through several modelling directions, from logistic regression using variable length Markov chains [to predict agreement between two of the three texts by regressing on earlier agreement] to hidden Markov models [representing, e.g., Matthew’s use of Mark], to various independence tests on contingency tables, sometimes bringing into the model an extra source denoted by Q. Including some R code for hidden Markov models. Once again, from my outsider viewpoint, this fragmented approach to the problem sounds problematic and inconclusive. And rather verbose in extensive discussions of descriptive statistics. Not that I was expecting a sudden Monty Python-like ray of light and booming voice to disclose the truth! Or that I crave for more p-values (some may be found hiding within the book). But I still wonder about the phylogeny… Especially since phylogenies are used in text authentication as pointed out to me by Robin Ryder for Chauncer’s [sic] Canterbury Tales.We can certainly list among the reasons for such omission the diffidence of the textual community towards phylogenetic methods, especially when performed by people from outside the field; but the potential reception problems for texts of enormous religious significance cannot be ruled out. However, one of the reasons might be far more trivial: the fact that, just as in the case of historical linguistics, we don’t have digital structured databases of the trove of data about this topic. Most of the literature is not even properly digital, at best with scanned PDFs. Furthermore, the data are usually far from perfect for such usage, as in the case of the synopsis by Smith (2017), which looks more like a typed table than a true database.
Next
In a future post, I will explore the problems of the synoptic gospels from a phylogenetic point of view, also releasing a minimal dataset (see The Synoptic Gospels problem: preparing a phylogenetic approach). Until then, those interested in the topic can find a lot of discussion on a mailing list devoted to the scholarly study of the synoptic gospels, Synoptic-L.
References
Abakuks, Andris (2014) The Synoptic Problem and Statistics. London: Chapman and Hall / CRC.
Goodacre, Mark (2001) The Synoptic Problem: a Way Through the Maze. New York: T & T Clark International. (available on Archive.org)
Robert, Christian P (2015) The synoptic problem and statistics [book review]. https://xianblog.wordpress.com/2015/03/20/the-synoptic-problem-and-statistics-book-review/
Orchard, Bernard; Longstaff, Thomas RW (1979) J.J. Griesbach: Synoptic and Text - Critical Studies 1776-1976. Cambridge: Cambridge University Press.
Smith, Mahlon H (2017) A Synoptic Gospels Primer. http://virtualreligion.net/primer/
Subscribe to:
Posts (Atom)















